Difference between revisions of "Template:Join paired-end FASTQ"
From SEED
(→Join paired-end FASTQ) |
(→Join paired-end FASTQ) |
||
| Line 1: | Line 1: | ||
==Join paired-end FASTQ== | ==Join paired-end FASTQ== | ||
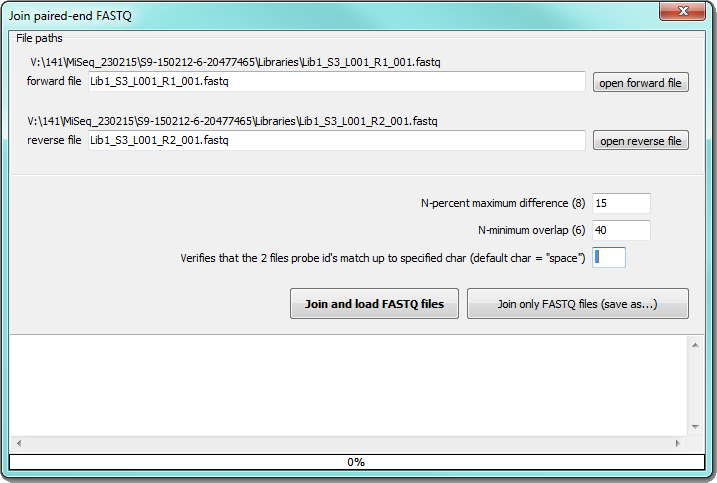
| − | To join pared-end reads from Illumina external program [[Settings|fastq-join]] is used. | + | To join pared-end reads from Illumina external program [[Settings|fastq-join]] is used. Define paths to forward and reverse files obtained from Illumina run, then you can select percentage of maximal difference and minimal overlap of the reads. The reads with the identical titles up to specified character (space is default) will be tested: |
{|class="wikitable" | {|class="wikitable" | ||
|- | |- | ||
Revision as of 12:45, 15 July 2015
Join paired-end FASTQ
To join pared-end reads from Illumina external program fastq-join is used. Define paths to forward and reverse files obtained from Illumina run, then you can select percentage of maximal difference and minimal overlap of the reads. The reads with the identical titles up to specified character (space is default) will be tested:
| from Lib1_S3_L001_R1_001.fastq | from Lib1_S3_L001_R2_001.fastq |
|---|---|
| M00816:79:000000000-AADTK:1:2115:18256:2080 1:N:0:3 | M00816:79:000000000-AADTK:1:2115:18256:2080 2:N:0:3 |
You can store joined and unpaired sequences in desired folder by "Join only FASTQ files (save as...)" button or you can load joined sequences to SEED by "Join and load FASTQ files" button.