Template:Program paths
Contents
Program paths
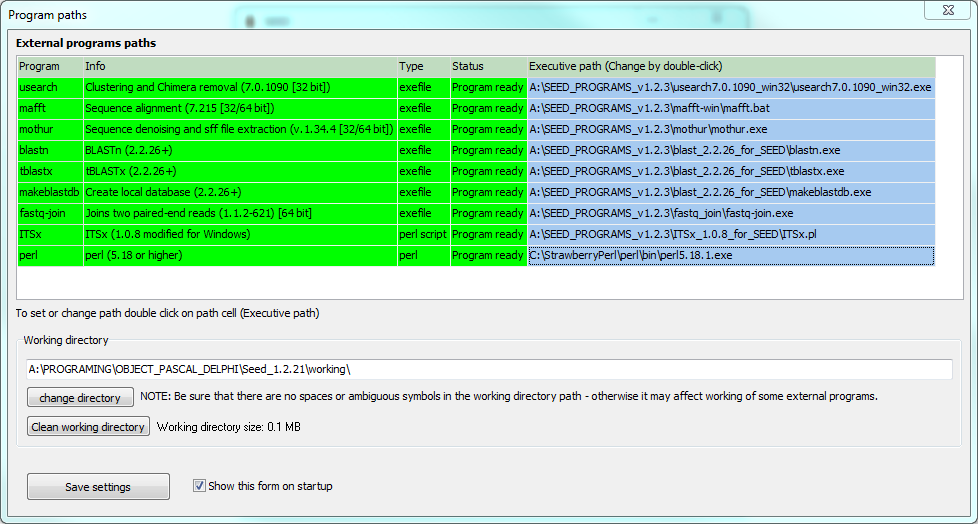
To run several functions you must set the program paths to the binaries of external programs. Paths can be set by double clicking the orange colored cell in appropriate row in external programs paths table.
When the path is set row color changes to green.
Please note: The green color does not mean that the path is correct - only means that it is set.
External programs
USEARCH
For correct functionality please use the appropriate version of the program (7.0.1090). This program enables clustering (UPARSE algorithm) and chimera removal (UCHIME algorithm).
Download: USEARCH
mafft
For correct functionality please use the appropriate version of the program (7.215). This program enables multiple sequence alignment.
Download: mafft
mothur
For the correct functionality please use the appropriate version of the program (1.34.4). This program enables sff file extraction and denoising. Note: For the denoising you must put LookUp_Titanium.pat file to
Download: mothur
BLAST TOOLS: blastn, tblastx and makeblastdb
These programs enable blasting against nucleotide databases (local or remote) and creating the local databases from your datasets (see more here). You can download Windows binary file here: blast_2.2.26_for_SEED.zip
FastqJoin
This program enables joining the pair-end reads from Illumina. You can download Windows binary file here: fastq_join.zip
ITSx
This program enables internal transcribed spacers extraction. You can download modified perl script for Windows (ITSx version 1.0.8) together with binaries of "hmmer" v3.0 here: ITSx_1.0.8_for_SEED.zip
Perl
To allow the SEED to run the perl script please select binary of instaled Strawberry Perl for Windows version 5.18 or higher.