
21st of June, 2025
Briefly but with full intensity! Kristina and Anya … should be called Dear Drs. from now on 🥳 – whohoooo! Kristina will join Hanka Hanzlikova’s lab (IMG CAS, Prague) as her new post-doc towards the end of 2025 (the best of luck!), Anya will stay to be gently tortured further in our lab (the best of luck too!) – whohoooo! New PhD student, Radu Centnerová, from Moravia will join the lab very soon, working closely with Anička – whohoooo! New Diploma student of Adri CZ, Bc. Mykhailo Streltsov from Ukraine, will join the lab very soon too – whohoooo! Peta and Zuzka are expecting babies – whohoooo! (I mean, not together! 😵💫) Klarka got married a week ago – whohoooo! Adri SK is no longer Leoš's PhD student, but newly Adri’s CZ - whohoooo! (I mean, what a shame! 🙃) Leoš has become a new member of the Academia Europaea association – whohoooo! (I mean, guys, who the … cares, right?!🤠) Leoš & comp advanced to the 3rd round of the ERC Synergy grant competition - whohoooo! (I mean, we ALL very MUCH care!!! 🫶 ) Filip CZ will soon be on his way to Trumpland to nail our future Nature paper down in Jodi Puglisi’s lab in Stanford – whohoooo! (I mean, the best of luck with everything in this current EverWonderLand!🤯) Pasha decided to leave us for India but recently changed his mind and will stay – whohoooo! (I mean, such a restless mind for his age ☺️.) NKOTB have gained a lot of skills and are now working with full force ✌️ – whohoooo! (I mean, it's sheer joy to see them growing into independent researchers!) Many thanks to Filip SK for taking care of our web 🙏 - whohoooo! Summer holidays are here - whohoooo 🌞! Howgh - whohoooo 🤪!

1st of April, 2025
This past wknd we went on a long-planned and once or twice postponed winter teambuilding adventure. Yes, it is spring already, but who cares. True, it would have been fun to see our “tropical” colleagues learning to ski (that was THE original idea), nonetheless, instead of snow&ice, we saw the awakening of nature – the white-flowering valley of the Zdobnice river, frogs laying zillion of eggs, singing birds inviting their partners to a dance of love, joyfully swimming fish (did not see any but they definitely must have been there!), people getting their gardens ready for a later harvest, slowly opening village pubs with rotten drinks from the last year (no kidding!), etc. … some of us even indulged in an invigorating/rejuvenating morning bath in the same river ... check out the photos.
Other than that, Kristína and Anya are getting ready to defend their desire to become PhDs, Adriana Jr. is getting ready to take the PhD state exam off her shoulders, and Terka’s followers and I are getting ready to write a big big paper :o) In the meantime, our collaborative papers in Nature, NSMB and mBio have been published (see below) and a Standard Czech Science Foundation grant has been - after an enforced last minute remake - submitted with low energy (endlessly) but high hopes (obviously)! What else? The world has gone completely mad, but we R staying sane!!! For as long as we can…
13th of January, 2025
Happy New Year to everyone – Peace on Earth above all! ☮️ We started 2025 with … indeed, grant reports, but also with this: https://www.nature.com/articles/s41594-024-01450-z ... awaited for a looooong time!🤝 Now, a short-lived joy and let’s move on! Soon another NSMB will be out (it is a real struggle to deal with their editorial office, constantly understaffed, it is No-Go for us for any future submissions to this journal 🙈), a Nature paper describing a new branch of the Integrated Stress Response pathway, to which Leos contributed to, will be most probably ‘accepted in principle’ any day now … cool 😃! Another collaborative paper with Jula Lukes and Zdenek Paris has just been submitted to PNAS, and another one with Marek Elias will be submitted to (most likely) NAR soon. With Christine Vogel, we are working on revisions of our start-stop (AUG-stop elements in the 5’ UTRs of mRNAs) paper for NAR, and a couple of other stories from our lab are shaping up into individual stories to be written up this year 💪. All in all, let’s wish that this year is a smooth ride through our research, as well as personal lives 🤘. I am very happy for sharing my life with you, guys! Thank you. 🙏 Leos
22st of November, 2024
Guess what? Zuzka’s and Petra’s Nature Structure & Molecular Biology paper has been accepted in principle after 17 super long months.🥳 Unbelievable! I mean, unbelievable that it took the editor over a year to just administer our manuscript. Everything was held up by one very negative reviewer, anonymous of course, some of whose comments were - based on cross-review by other reviewers - ridiculous and for whatever reason hostile, and who essentially took the position of the editor, with the real editor playing only the role of secretary... I consider this to be a major failure of the editor's job! 🙈
Never mind, if this negative reviewer ever reads these lines, I encourage them to contact me, so that we can talk about anything they didn’t like. I’ll try to explain it to “you” and will listen to your thoughts, because “constructive criticism with a face” is much easier to deal with than "mean criticism" from someone steadfastly hiding in the darkness of anonymity. But all is well that ends well. After filing a complaint with the Editor-in-Chief of the entire Nature group, we got a new editor, who finally accepted it and apologized deeply for all the time lost. Chief Editor of NSMB also apologized profusely, and told us that she really liked our work very much, so she will highlight it with a News & Views commentary. 🤠
All in all, big congratulations girls, to Zuzka for her fantastic work at the beginning and to Petra for her fantastic work at the end. The middle was in common! 😎🤙
In addition to that, one more collaborative NSMB paper where Leos is a co-corresponding author was accepted in principle on the same day (led by Pasha Baranov and Gary Loughran)! 😇 A Communication Biology collaborative paper with a Japanese group of Takeshi Chujo was also published this month, and finally Anya’s Star Protocols paper was published just yesterday 🥳. So at the beginning of this month our thoughts were as dark as the sky over Prague🌧️, but now they are as full of sunshine as the sky over Prague. 🌞
1st of Oct, 2024
TITLE: A proloooonged weekend in the heart of western Šumava opened our hearts once and for all.
ABSTRACT: People enjoy having fun. However, people have to go to work (almost) every day to work very hard, so that there is practically no time for socializing. What to do then? To fill this critical knowledge gap and examine the effect of being together day & night as a team for almost 3 days, we took advantage of our private road devices (so-called cars) and went to the western Šumava region near Kdyně - Penzion Pranty. We tested hiking, magic mushroom (Bedlas Jedlas) picking & frying, meat & vegan grilling, Indian cooking, overeating & overdrinking, singing, playing volleyball, ping-pong, billiard, swimming :) … simply explored how to have fun as a team. All activities scored positive on our pleasure scale with p < 0.000001. Truly, it was fun 😎. In detail, on Friday we peaked on Paradise and got drunk, on Saturday we hiked the highest peak of this region - Čerchov (a super steep and long and exhausting hike 🤪) and got drunk, and on Sunday we almost went horseback riding - instead we ended up in the pilgrimage church of St. Anna on Tanaberk and prayed. The whole weekend experience had a very sunny impact on our souls, not so sunny on our stomachs & heads, but that was overridden by colorful and tasty headache pills. And the fabulous Sunday weather too.
Concluding remarks: The take home message of this common experiment is very straightforward: let’s do it again soon!
Funding: Endless Universe 🙏.
Acknowledgments: To all who joined, as well as to all that did not (Endless Universe could save some money 🥳).
References: booking.com, mapy.cz, whatsApp and instagram (at least for some of us 😉)
2nd of Sept, 2024
Kids are back at school, hhuurraayy! 😁 Petra is about to take her state exam and defend her diploma work and Srinjoyee has finally joined the NKOTB club. This means that we R complete & ready to roll! 🤙Our team-building weekend in the Sumava mountain is drawing near! 🥳 What else? It is hot hot hot!!! 😅
22th of July, 2024
Kristina’s Tma manu has just been accepted in Communication Biology from the Nature Portfolio publishing group! 🤯🥳 Big congrats and many thanks for your practically independent work 👍👍! With this accomplishment, Kristina's path to her PhD degree is clear and leads in only one direction ... to nail it down soon 💪Fingers crossed! 🤙
15th of July, 2024
Guess what? Nawal, our new PhD student from Lebanon, has joined the lab (and brought us an excellent assortment of baklava and cones of mystical Cedrus libani with a very pleasant scent) and Leos was elected to the EMBO Membership. Most importantly, Terka is organizing a summer barbecue at the end of July before leaving us for her eagerly awaited maternity duties 😁
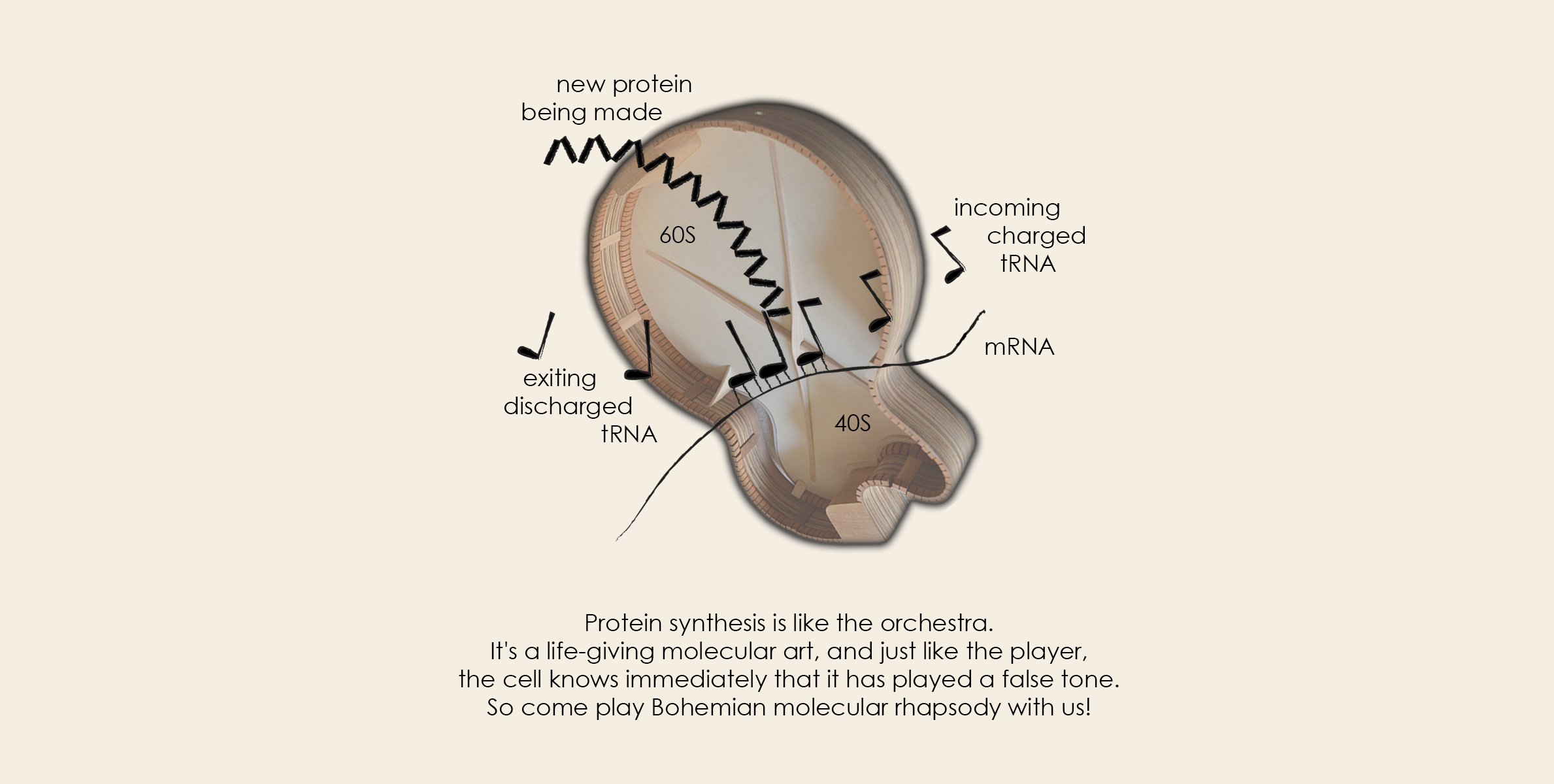
We are a medium-sized group (~15 people) of enthusiastic, passionate and hard-working “musicians” from post-docs to undergraduate students with one skillful lab assistant, who are eager to learn more and more every day while having fun at the same time. Our recording studio is based at the Institute of Microbiology (IMIC) of the Czech Academy of Sciences (CAS) in Prague, Czech Republic.
Our band nicknamed Laboratory of Regulation of Gene Expression (LRGE) was established in June 2006 and investigates fundamentals of one of the key, life-giving molecular processes in the cell and its regulation at both general as well as gene-specific levels. We R talking about protein synthesis! Simply speaking, our mostly basic research is focused on figuring out how the cell deciphers the genetic code of DNA using relatively simple decoding keys and uses this gene-encoded information to produce proteins, i.e., cellular effectors, through a very complex process also called translation.
Recently, we have become very much interested in various alternatives to these decoding keys, so-called alternative genetic codes, the detailed knowledge of which could be exploited in the treatment of various genetic diseases. In addition, we are interested in understanding how translation is regulated in response to constantly changing environmental conditions that the cell faces and to which it must immediately respond in order to adapt its repertoire of proteins that it is currently producing.


As for the importance of our research from the socio-economic and medical perspectives, we want to broaden and deepen the general understanding of how these basic cellular processes properly operate in the cell, so that our collaborators who are primarily interested in applied research; i.e., research directly aimed at developing the most appropriate treatment for a particular disease, can feed from it while investigating various pathologies.
Overall, paying the utmost tribute to our distinguished ancient predecessors, our major goal is to help to transmute lead ( basic research discoveries) into gold (useful therapeutics) with the simplest and finest tones.
Our compositions combine the use of mammalian cells lines and partly also the budding yeast Saccharomyces cerevisiae model organism and employ tools of molecular and structural biology, biochemistry and genetics.
We also very much enjoy participating in numerous popularization gigs (like Open Science, Researcher’s Night, etc.), giving interviews to news and radio, contributing to popularization magazines, etc.
Our band won “The best original research publication carried out at the Institute of Microbiology CAS Award” (our home institute) in years 2008, 2011, 2013, 2015, 2016, 2017, 2018, 2019, 2020, 2021, 2022 and 2023. In February 2016, the international evaluation committee selected our band as one of the 13 most prestigious ones in the entire CAS.
Our recording is (or until recently was) supported by the national and world renowned funding bodies such as the the Wellcome Trust, Howard Hughes Medical Institute, NIH FIC, German Science Foundation, Czech Science Foundation and Czech Academy of Sciences (Praemium Academiae). Altogether, we have produced >80 records with record labels like Nature (see our winning cover page), Molecular Cell, Nature Structural & Molecular Biology, Nature Protocols, Nucleic Acids Research, Genes & Development, EMBO J., eLife, etc. Music is always composed by LRGE, lyrics usually by LSV.





Group leader (EMBO member)
e-mail: valasekl@biomed.cas.cz

Independent project leader
e-mail: adriana.roithova@biomed.cas.cz

Research scientist - lab manager
e-mail: herrmannova@biomed.cas.cz

Research scientist
e-mail: mpasha@biomed.cas.cz

Research scientist
e-mail: jana.chumova@biomed.cas.cz
Research scientist
e-mail: jana.vojtova@biomed.cas.cz

Post-doctoral fellow (maternity leave)
e-mail: vladislava.hronova@biomed.cas.cz

Post-doctoral fellow
e-mail: filip.brazdovic@biomed.cas.cz

Post-doctoral fellow
e-mail: filip.trcka@biomed.cas.cz

Post-doctoral fellow (maternity leave)
e-mail: zuzana.pavlikova@biomed.cas.cz

Post-doctoral fellow (maternity leave)
e-mail: terezie.svobodova@biomed.cas.cz

Post-doctoral fellow
e-mail: anna.smirnova@biomed.cas.cz

Post-doctoral fellow
e-mail: kristina.jendruchova@biomed.cas.cz

Ph.D. student
e-mail: klara.pospisilova@biomed.cas.cz

Ph.D. student
e-mail: adriana.subrtova@biomed.cas.cz
Ph.D. student
e-mail: petra.miletinova@biomed.cas.cz

Ph.D. student
e-mail: nawal.alchamy@biomed.cas.cz
Ph.D. student
e-mail: pragya.kamal@biomed.cas.cz

Ph.D. student
e-mail: srinjoyee.pawar@biomed.cas.cz

Ph.D. student
e-mail: to_be_announced@biomed.cas.cz

Diploma student
e-mail: to_be_announced@biomed.cas.cz

Lab assistant
e-mail: olga.krydova@biomed.cas.cz
Institute of Microbiology, AS CR, v.v.i.
Vídeňská 1083, Prague 4, 142 20
Czech Republic
Phone: +420 241 062 288 (2483)
Cell: +420 724 731 661
e-mail: valasekl@biomed.cas.cz
Take C-line metro to the station „Kačerov“.
Get on the bus 138 to the final station "Ústavy akademie věd".
We are in the Bldg. L, second floor, lab #128.

created by DALL·E 3
Guess the song:
Pa papa pa papapapa pa, pa papa pa papapapa pa, pa pa papapapapa pa, papapapapa pa... what is it? :o)
Guess the songs:
🔇🔈🔉🔊
👁️🐅
☀️🏖️ 6️⃣9️⃣
☔ 👨🌧️👨